Placing volumes that contain multiple DNA solenoids
Multiple solenoid volumes allow for the densest DNA geometries we generate here.
Up to 9 solenoids can be placed in a voxel by the methods shown in this example.
It is recommended that you run this example inside a Jupyter environment rather than a VSCode or similar environment
This requires the mayavi jupyter extension jupyter nbextension install --py mayavi --user
[ ]:
import sys
from pathlib import Path
try:
from fractaldna.dna_models import dnachain as dna
except (ImportError, ModuleNotFoundError):
sys.path.append(str(Path.cwd().parent.parent.parent))
from fractaldna.dna_models import dnachain as dna
from mayavi import mlab
# Disable this option for interactive rendering
mlab.options.offscreen = True
# Enable this option for an interactive notebook
# mlab.init_notebook()
Making Multiple Solenoids
[ ]:
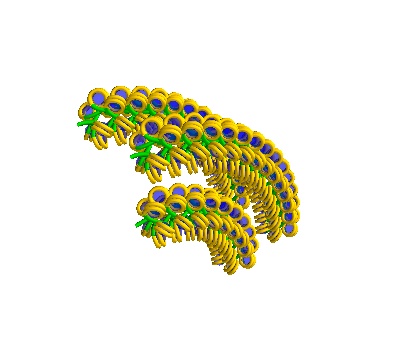
# Multi-Solenoidal Volumes contain 8 chromatin strands.
chain = dna.MultiSolenoidVolume(voxelheight=750)
# MayaVI plots are best for visualisation here
plot = chain.to_strand_plot()
plot.scene.save_jpg("multi_solenoid_strand_plot.jpg")

[ ]:
# They can be turned, and only select chains can be generated
chain = dna.MultiSolenoidVolume(
voxelheight=1500, separation=400, turn=True, chains=[0, 3, 7]
)
plot = chain.to_line_plot()
plot.scene.save_jpg("multi_solenoid_turned_plot.jpg")
Exporting multiple solenoids as text
[ ]:
# This too can be exported to a dataframe of basepairs
chain.to_frame()
[ ]:
# And a second base pair of histones
chain.histones_to_frame()
[ ]: